Visualizing Uncertainty
2025-06-22
We all know how to visualize uncertainty, right?
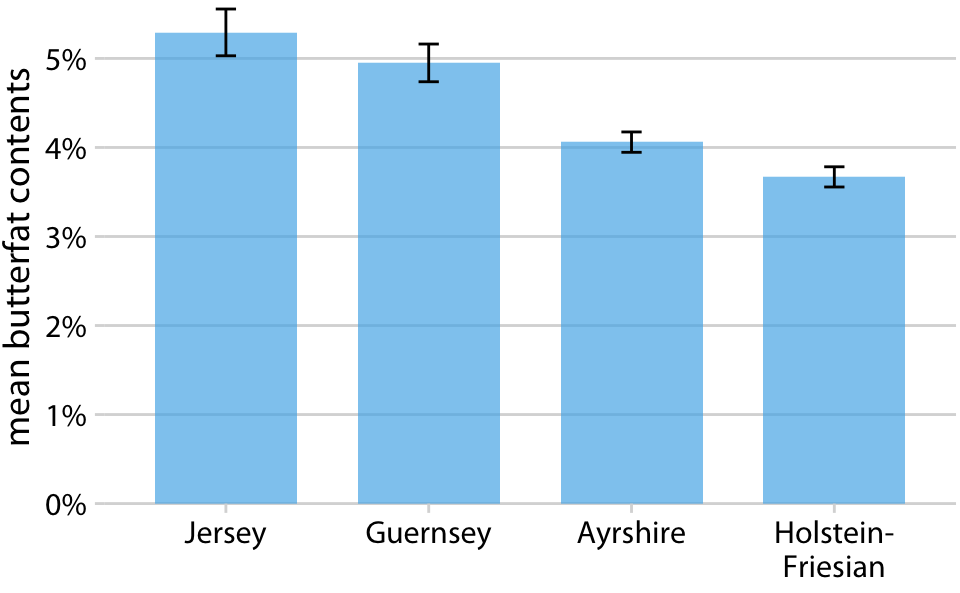
Milk butterfat contents by cattle breed. Source: Canadian Record of Performance for Purebred Dairy Cattle
We all know how to visualize uncertainty, right?
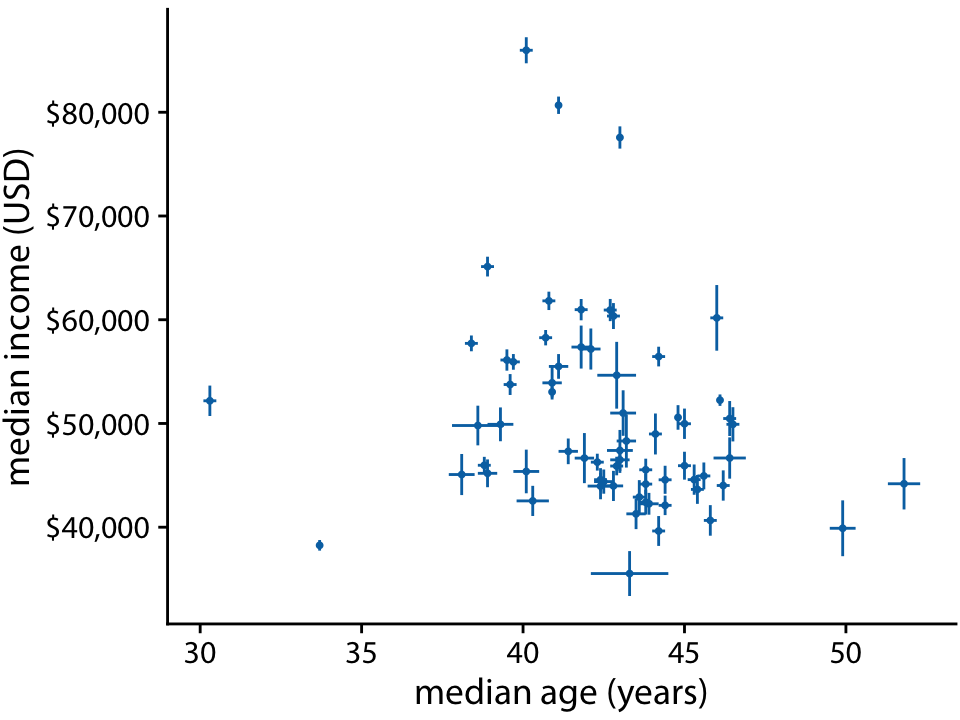
Income versus age for 67 counties in Pennsylvania. Source: 2015 Five-Year American Community Survey
We all know how to visualize uncertainty, right?
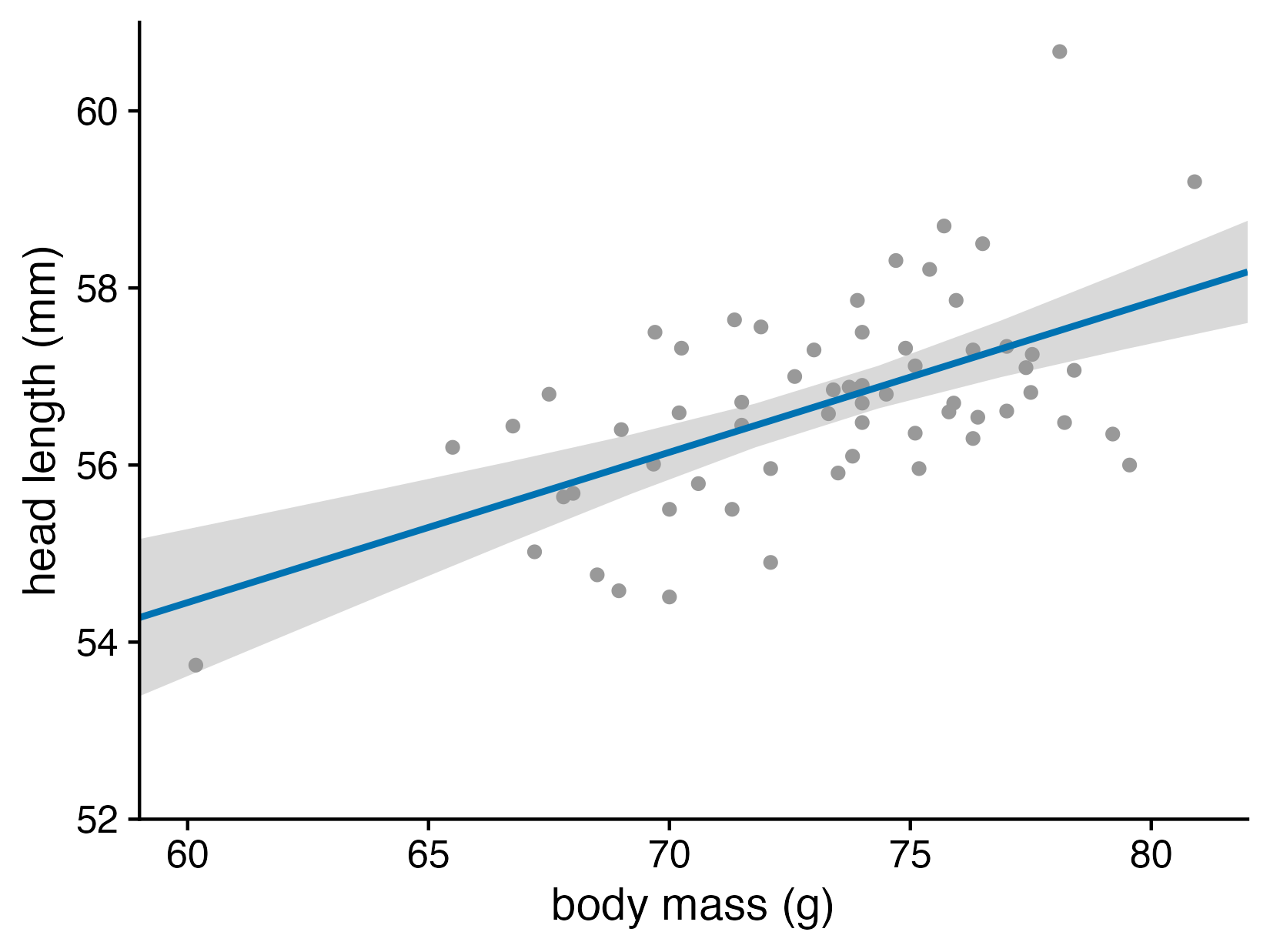
Head length versus body mass for male blue jays. Data source: Keith Tarvin, Oberlin College
These commonly used visualizations have problems
- It’s often not clear what exactly the visualizations represent
- Even if we know, we can have difficulty interpreting what we see
It’s often not clear what the visualizations represent
In particular, error bars can represent many different quantities
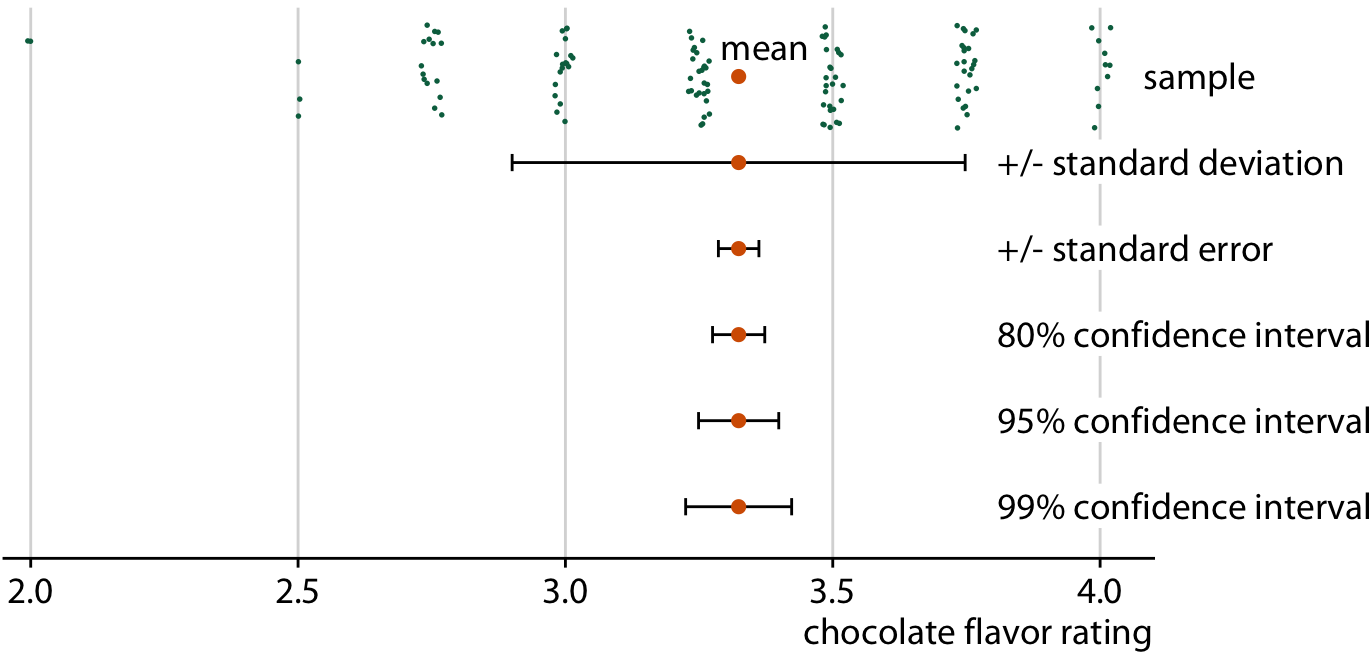
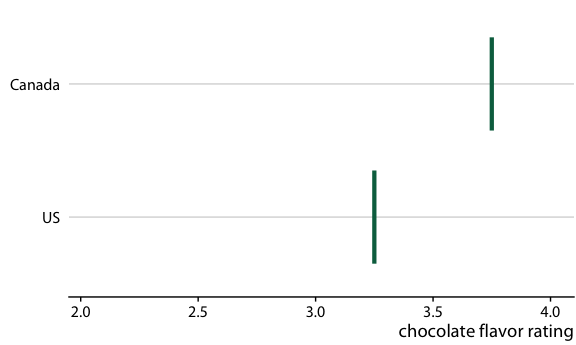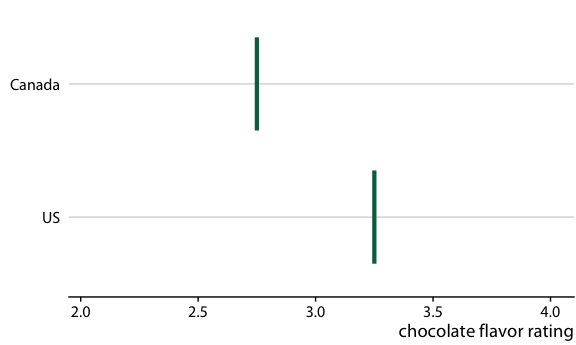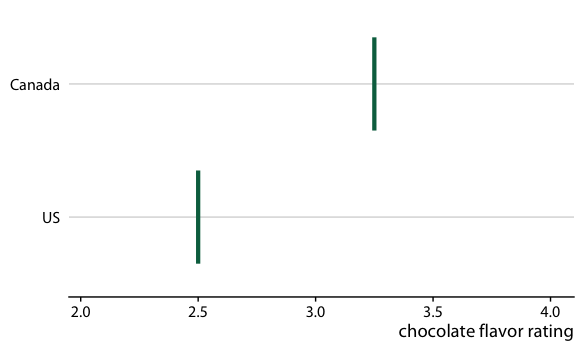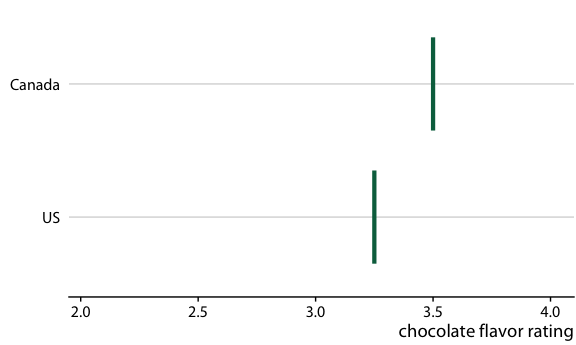
Chocolate bar ratings. Source: Manhattan Chocolate Society
We can have difficulty interpreting what we see
- People are not good at reasoning about probabilities
- People will substitute uncertainty with simpler concepts
(Deterministic Construal Error) - People will interpret intervals as hard boundaries
Lace Padilla, Matthew Kay, and Jessica Hullman (2022): Uncertainty Visualization
Uncertainty shown as 95% CI error bars with caps
Chocolate bars from four countries compared to bars from the US
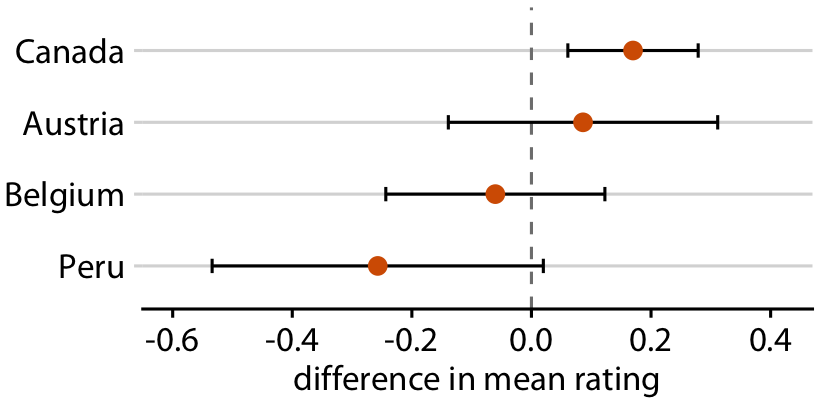
Relative rankings compared to US chocolate bars. Source: Manhattan Chocolate Society
Uncertainty shown as 95% CI error bars with caps
Determinstic Construal Error:
Error bars are interpreted as min/max values

Relative rankings compared to US chocolate bars. Source: Manhattan Chocolate Society
Uncertainty shown as 95% CI error bars with caps
Categorical thinking:
Areas outside and inside the error bars are categorically different

Relative rankings compared to US chocolate bars. Source: Manhattan Chocolate Society
Uncertainty shown as 95% CI error bars without caps
You can remove caps to make the boundary visually less severe
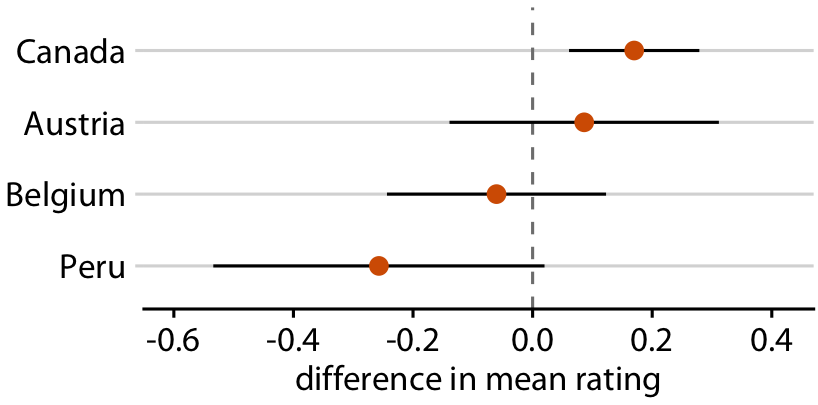
Relative rankings compared to US chocolate bars. Source: Manhattan Chocolate Society
Uncertainty shown as graded error bars
You can show multiple confidence levels to de-emphasize existence of boundary
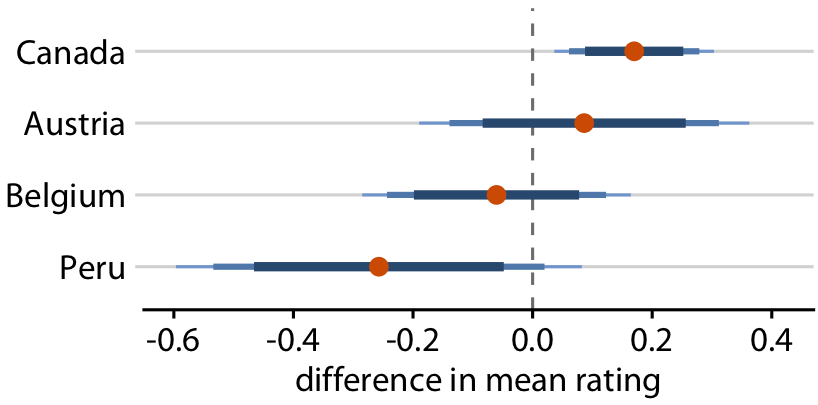
Relative rankings compared to US chocolate bars. Source: Manhattan Chocolate Society
Uncertainty shown as confidence strips
You can use faded strips (but hard to read/interpret)
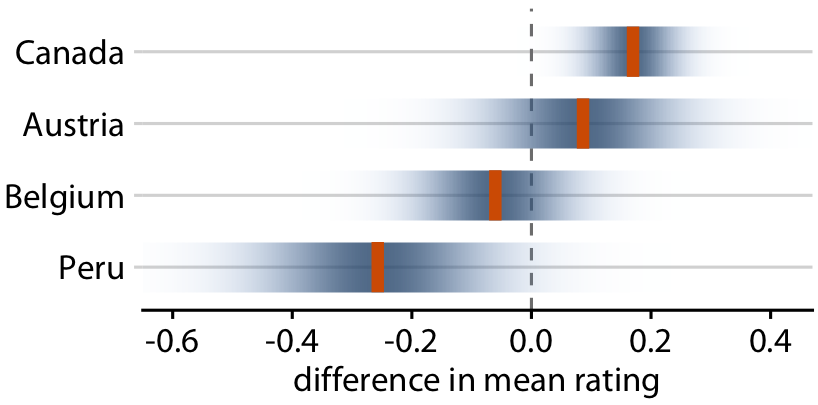
Relative rankings compared to US chocolate bars. Source: Manhattan Chocolate Society
Uncertainty shown as distributions
You can show actual distributions
Popular in Bayesian inference, but still difficult to interpret
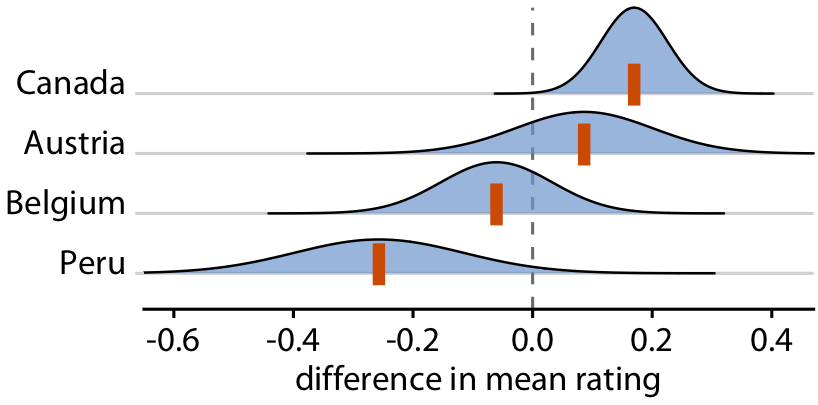
Relative rankings compared to US chocolate bars. Source: Manhattan Chocolate Society
Consider the actual experience of a customer
If I can buy either a Canadian or a US bar, what is the probability that the Canadian bar will be better?
Answer: The Canadian bar has a 53% chance of being better
How can we communicate this?
Use frequency framing to communicate probabilities
Use frequency framing to communicate probabilities
Use frequency framing to communicate probabilities
Use frequency framing to communicate probabilities
Use frequency framing to communicate probabilities
Alternatively: Use Hypothetical Outcome Plots
Hypothetical Outcome Plots use animation to let viewers experience uncertainty
Alternatively: Use Hypothetical Outcome Plots
Hypothetical Outcome Plots use animation to let viewers experience uncertainty
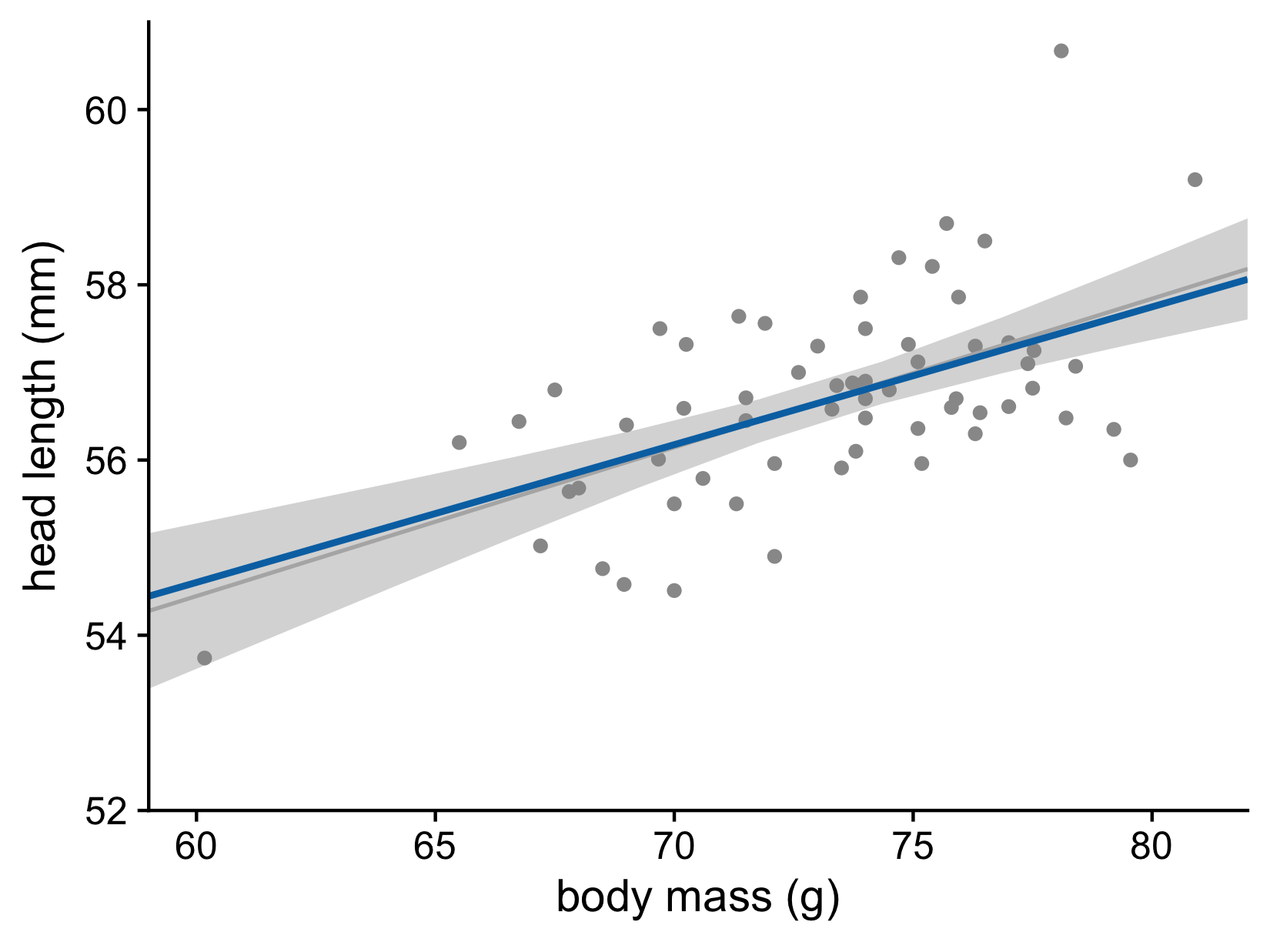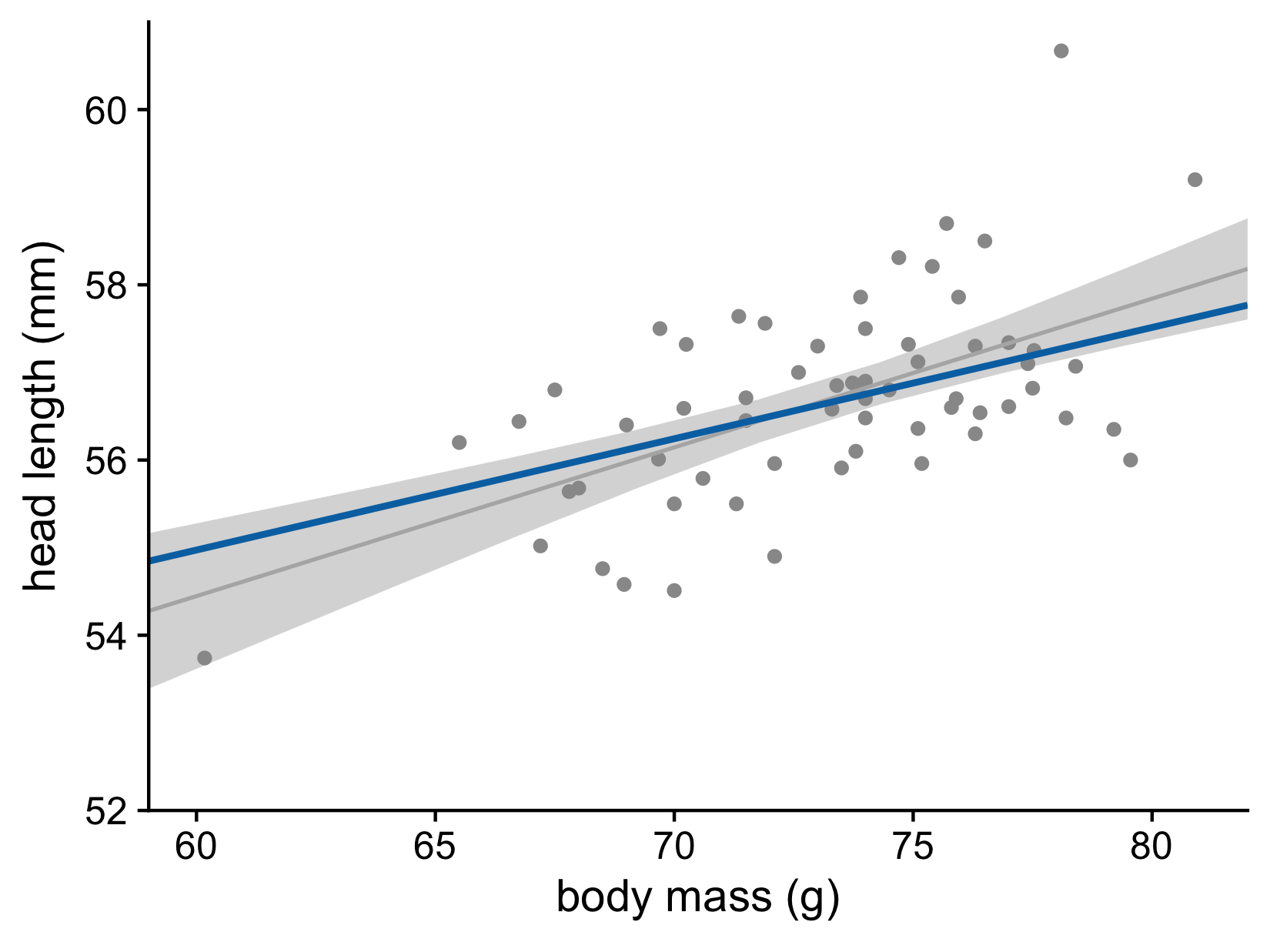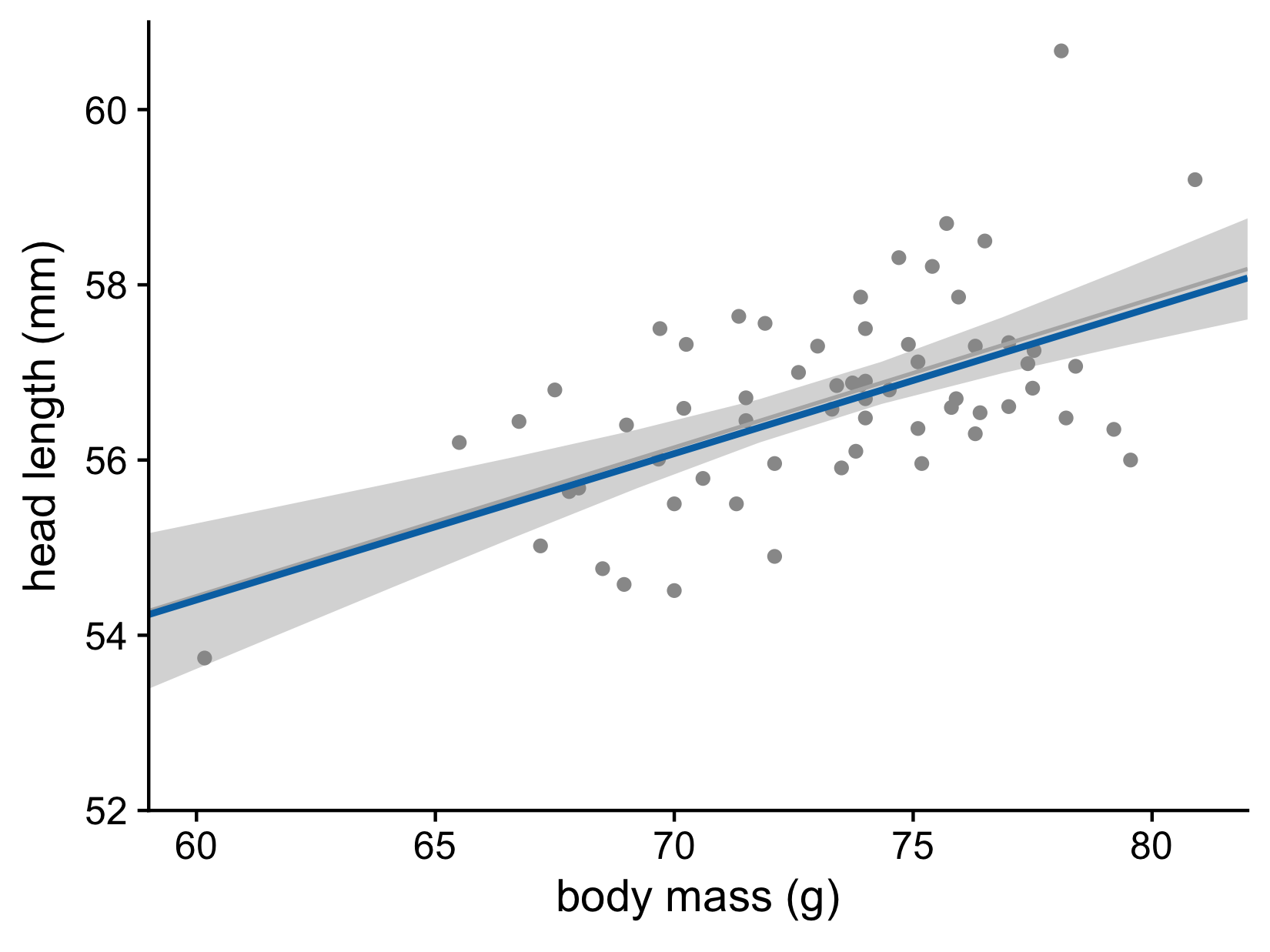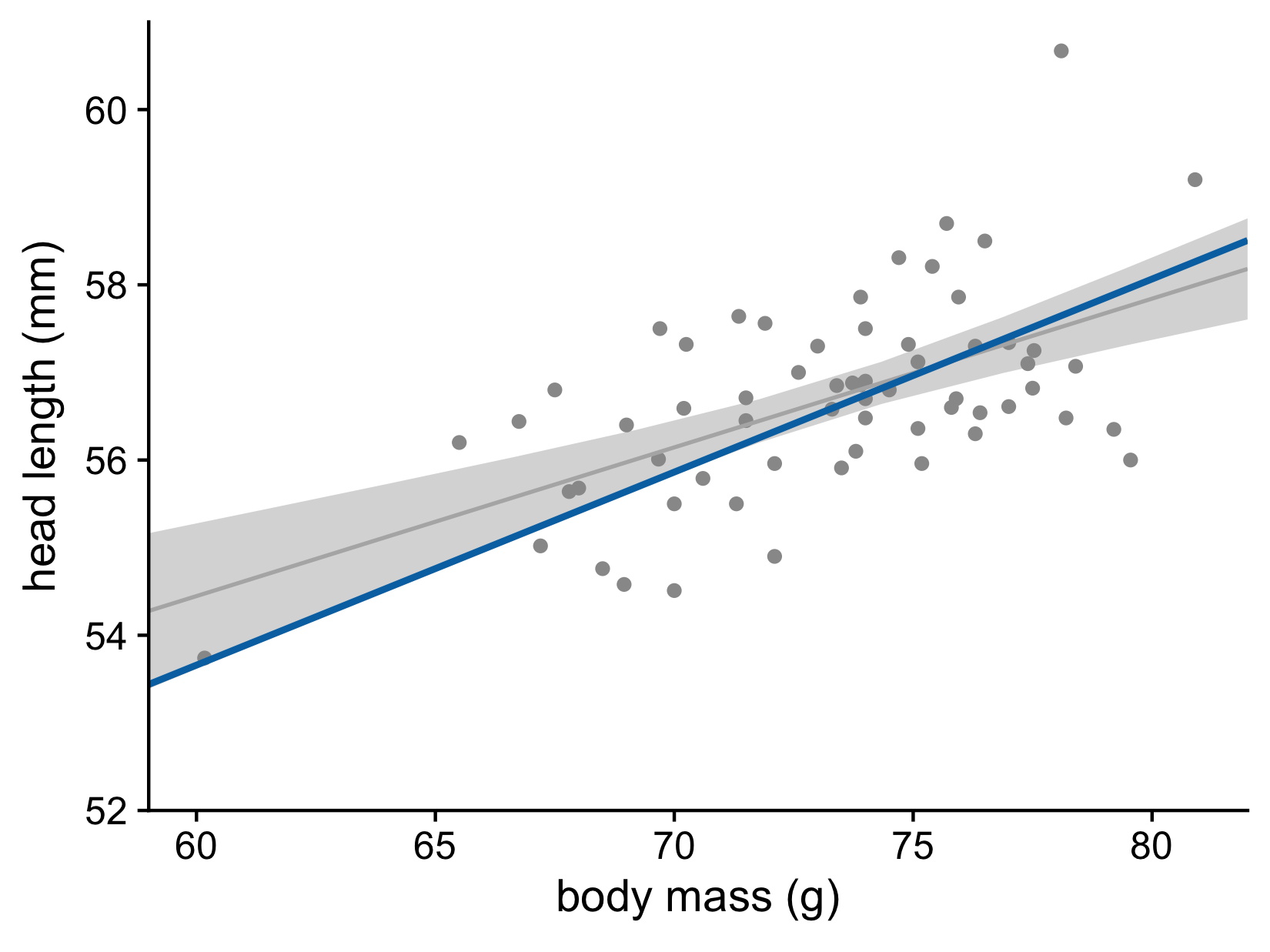
Let’s now consider the uncertainty of trend lines
What does the confidence band show?

Head length versus body mass for male blue jays. Data source: Keith Tarvin, Oberlin College
Let’s now consider the uncertainty of trend lines
Both the intercept and the slope have uncertainty
Head length versus body mass for male blue jays. Data source: Keith Tarvin, Oberlin College
It gets even more confusing for non-linear trend lines
Individual sample fits tend to be more wiggly than the mean
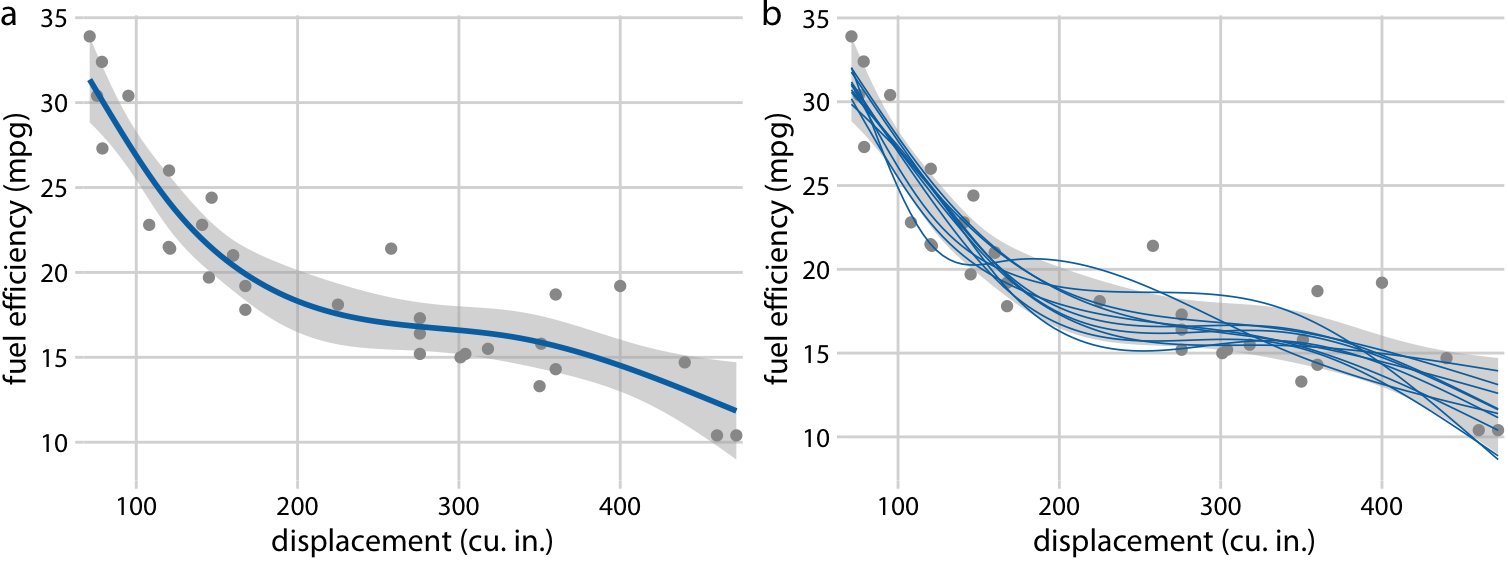
Fuel efficiency versus displacement, for 32 cars (1973–74 models). Source: Motor Trend, 1974
Hypothetical Outcome Plots again help develop intuition
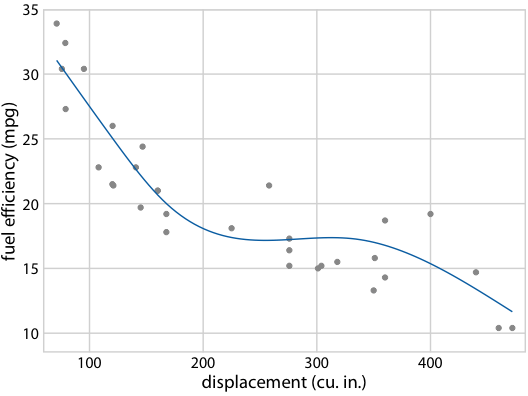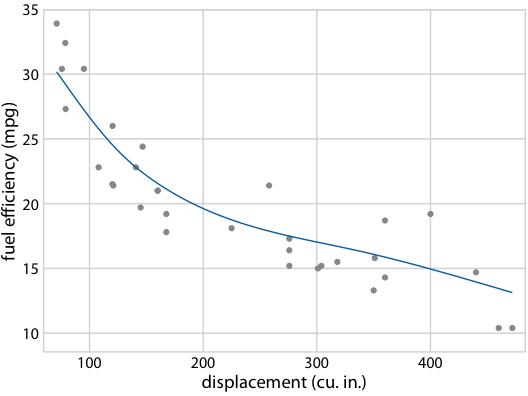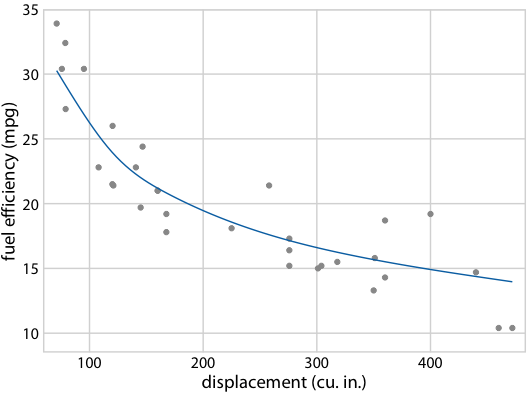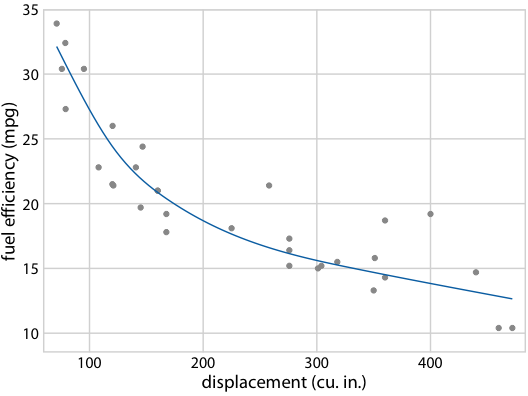
Fuel efficiency versus displacement, for 32 cars (1973–74 models). Source: Motor Trend, 1974
Uncertainty visualizations in R
Getting the data
The gapminder dataset:
# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsMaking a plot with error bars
Example: Relationship between life expectancy and GDP per capita
Making a plot with error bars
Example: Relationship between life expectancy and GDP per capita
Making a plot with error bars
# A tibble: 60 × 3
continent year data
<fct> <int> <list>
1 Asia 1952 <tibble [33 × 4]>
2 Asia 1957 <tibble [33 × 4]>
3 Asia 1962 <tibble [33 × 4]>
4 Asia 1967 <tibble [33 × 4]>
5 Asia 1972 <tibble [33 × 4]>
6 Asia 1977 <tibble [33 × 4]>
7 Asia 1982 <tibble [33 × 4]>
8 Asia 1987 <tibble [33 × 4]>
9 Asia 1992 <tibble [33 × 4]>
10 Asia 1997 <tibble [33 × 4]>
# ℹ 50 more rowsMaking a plot with error bars
lm_data <- gapminder |>
nest(data = -c(continent, year)) |>
mutate(
fit = map(data, ~lm(lifeExp ~ log(gdpPercap), data = .x))
)
lm_data# A tibble: 60 × 4
continent year data fit
<fct> <int> <list> <list>
1 Asia 1952 <tibble [33 × 4]> <lm>
2 Asia 1957 <tibble [33 × 4]> <lm>
3 Asia 1962 <tibble [33 × 4]> <lm>
4 Asia 1967 <tibble [33 × 4]> <lm>
5 Asia 1972 <tibble [33 × 4]> <lm>
6 Asia 1977 <tibble [33 × 4]> <lm>
7 Asia 1982 <tibble [33 × 4]> <lm>
8 Asia 1987 <tibble [33 × 4]> <lm>
9 Asia 1992 <tibble [33 × 4]> <lm>
10 Asia 1997 <tibble [33 × 4]> <lm>
# ℹ 50 more rowsMaking a plot with error bars
lm_data <- gapminder |>
nest(data = -c(continent, year)) |>
mutate(
fit = map(data, ~lm(lifeExp ~ log(gdpPercap), data = .x)),
tidy_out = map(fit, tidy)
)
lm_data# A tibble: 60 × 5
continent year data fit tidy_out
<fct> <int> <list> <list> <list>
1 Asia 1952 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
2 Asia 1957 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
3 Asia 1962 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
4 Asia 1967 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
5 Asia 1972 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
6 Asia 1977 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
7 Asia 1982 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
8 Asia 1987 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
9 Asia 1992 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
10 Asia 1997 <tibble [33 × 4]> <lm> <tibble [2 × 5]>
# ℹ 50 more rowsMaking a plot with error bars
lm_data <- gapminder |>
nest(data = -c(continent, year)) |>
mutate(
fit = map(data, ~lm(lifeExp ~ log(gdpPercap), data = .x)),
tidy_out = map(fit, tidy)
) |>
unnest(cols = tidy_out)
lm_data# A tibble: 120 × 9
continent year data fit term estimate std.error statistic p.value
<fct> <int> <list> <list> <chr> <dbl> <dbl> <dbl> <dbl>
1 Asia 1952 <tibble> <lm> (Interc… 15.8 9.27 1.71 9.78e-2
2 Asia 1952 <tibble> <lm> log(gdp… 4.16 1.25 3.33 2.28e-3
3 Asia 1957 <tibble> <lm> (Interc… 18.1 9.70 1.86 7.20e-2
4 Asia 1957 <tibble> <lm> log(gdp… 4.17 1.28 3.26 2.71e-3
5 Asia 1962 <tibble> <lm> (Interc… 16.6 9.52 1.74 9.11e-2
6 Asia 1962 <tibble> <lm> log(gdp… 4.59 1.24 3.72 7.94e-4
7 Asia 1967 <tibble> <lm> (Interc… 19.8 9.05 2.19 3.64e-2
8 Asia 1967 <tibble> <lm> log(gdp… 4.50 1.15 3.90 4.77e-4
9 Asia 1972 <tibble> <lm> (Interc… 21.9 8.14 2.69 1.13e-2
10 Asia 1972 <tibble> <lm> log(gdp… 4.44 1.01 4.41 1.16e-4
# ℹ 110 more rowsMaking a plot with error bars
lm_data <- gapminder |>
nest(data = -c(continent, year)) |>
mutate(
fit = map(data, ~lm(lifeExp ~ log(gdpPercap), data = .x)),
tidy_out = map(fit, tidy)
) |>
unnest(cols = tidy_out) |>
select(-fit, -data)
lm_data# A tibble: 120 × 7
continent year term estimate std.error statistic p.value
<fct> <int> <chr> <dbl> <dbl> <dbl> <dbl>
1 Asia 1952 (Intercept) 15.8 9.27 1.71 0.0978
2 Asia 1952 log(gdpPercap) 4.16 1.25 3.33 0.00228
3 Asia 1957 (Intercept) 18.1 9.70 1.86 0.0720
4 Asia 1957 log(gdpPercap) 4.17 1.28 3.26 0.00271
5 Asia 1962 (Intercept) 16.6 9.52 1.74 0.0911
6 Asia 1962 log(gdpPercap) 4.59 1.24 3.72 0.000794
7 Asia 1967 (Intercept) 19.8 9.05 2.19 0.0364
8 Asia 1967 log(gdpPercap) 4.50 1.15 3.90 0.000477
9 Asia 1972 (Intercept) 21.9 8.14 2.69 0.0113
10 Asia 1972 log(gdpPercap) 4.44 1.01 4.41 0.000116
# ℹ 110 more rowsMaking a plot with error bars
lm_data <- gapminder |>
nest(data = -c(continent, year)) |>
mutate(
fit = map(data, ~lm(lifeExp ~ log(gdpPercap), data = .x)),
tidy_out = map(fit, tidy)
) |>
unnest(cols = tidy_out) |>
select(-fit, -data) |>
filter(term != "(Intercept)", continent != "Oceania")
lm_data# A tibble: 48 × 7
continent year term estimate std.error statistic p.value
<fct> <int> <chr> <dbl> <dbl> <dbl> <dbl>
1 Asia 1952 log(gdpPercap) 4.16 1.25 3.33 0.00228
2 Asia 1957 log(gdpPercap) 4.17 1.28 3.26 0.00271
3 Asia 1962 log(gdpPercap) 4.59 1.24 3.72 0.000794
4 Asia 1967 log(gdpPercap) 4.50 1.15 3.90 0.000477
5 Asia 1972 log(gdpPercap) 4.44 1.01 4.41 0.000116
6 Asia 1977 log(gdpPercap) 4.87 1.03 4.75 0.0000442
7 Asia 1982 log(gdpPercap) 4.78 0.852 5.61 0.00000377
8 Asia 1987 log(gdpPercap) 5.17 0.727 7.12 0.0000000531
9 Asia 1992 log(gdpPercap) 5.09 0.649 7.84 0.00000000760
10 Asia 1997 log(gdpPercap) 5.11 0.628 8.15 0.00000000335
# ℹ 38 more rowsMaking a plot with error bars
Half-eyes, gradient intervals, etc
The ggdist package provides many different visualizations of uncertainty
Half-eyes, gradient intervals, etc
The ggdist package provides many different visualizations of uncertainty
library(ggdist)
library(distributional) # for dist_normal()
lm_data |>
filter(year == 1952) |>
mutate(
continent =
fct_reorder(continent, estimate)
) |>
ggplot(aes(x = estimate, y = continent)) +
stat_dist_gradientinterval(
aes(dist = dist_normal(
mu = estimate, sigma = std.error
)),
point_size = 4,
fill = "skyblue"
)Half-eyes, gradient intervals, etc
The ggdist package provides many different visualizations of uncertainty
library(ggdist)
library(distributional) # for dist_normal()
lm_data |>
filter(year == 1952) |>
mutate(
continent =
fct_reorder(continent, estimate)
) |>
ggplot(aes(x = estimate, y = continent)) +
stat_dist_dotsinterval(
aes(dist = dist_normal(
mu = estimate, sigma = std.error
)),
point_size = 4,
fill = "skyblue",
quantiles = 20
)Half-eyes, gradient intervals, etc
The ggdist package provides many different visualizations of uncertainty
library(ggdist)
library(distributional) # for dist_normal()
lm_data |>
filter(year == 1952) |>
mutate(
continent =
fct_reorder(continent, estimate)
) |>
ggplot(aes(x = estimate, y = continent)) +
stat_dist_dotsinterval(
aes(dist = dist_normal(
mu = estimate, sigma = std.error
)),
point_size = 4,
fill = "skyblue",
quantiles = 10
)Further reading
- Fundamentals of Data Visualization: Chapter 16: Visualizing uncertainty
- Data Visualization—A Practical Introduction: Chapter 6.6: Grouped analysis and list columns
- Data Visualization—A Practical Introduction: Chapter 6.7: Plot marginal effects
- ggdist reference documentation: https://mjskay.github.io/ggdist/index.html
- ggdist vignette: Frequentist uncertainty visualization