library(tidyverse)
library(colorspace)
library(cowplot)Effective Data Visualization with ggplot2
Sprucing up your legends, solutions to exercises
Load required packages
Solutions, Section 1
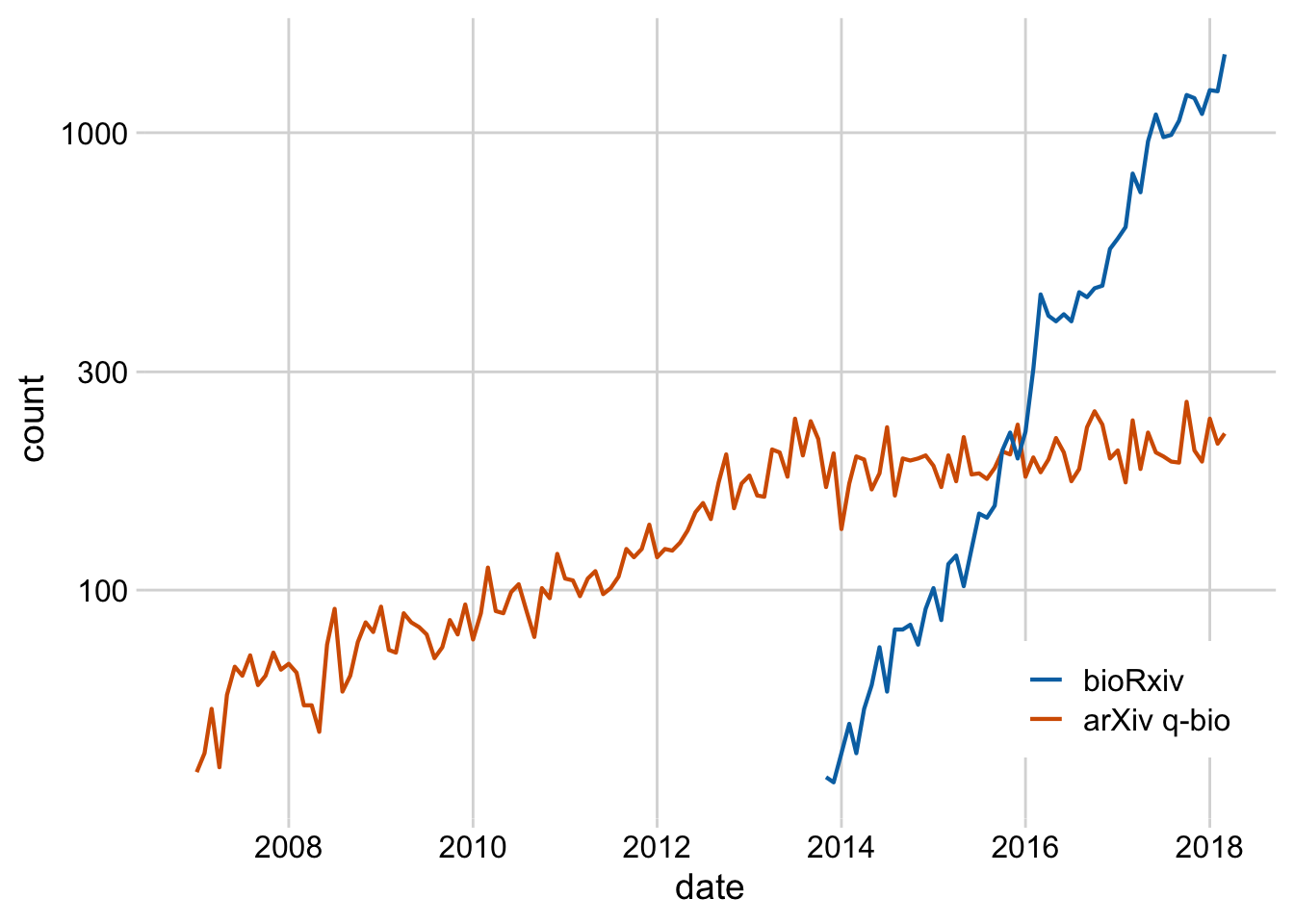
Exercise 1.1: Replace the legend in this plot with a secondary axis. Also style the plot.
preprints <- read_csv("https://wilkelab.org/dataviz_shortcourse/datasets/preprints.csv") |>
filter(archive %in% c("bioRxiv", "arXiv q-bio")) |>
filter(count > 0)
ggplot(preprints) +
aes(date, count, color = archive) +
geom_line(linewidth = 0.75) +
scale_y_log10() +
scale_x_date() +
scale_color_manual(values = c("#D55E00", "#0072B2"))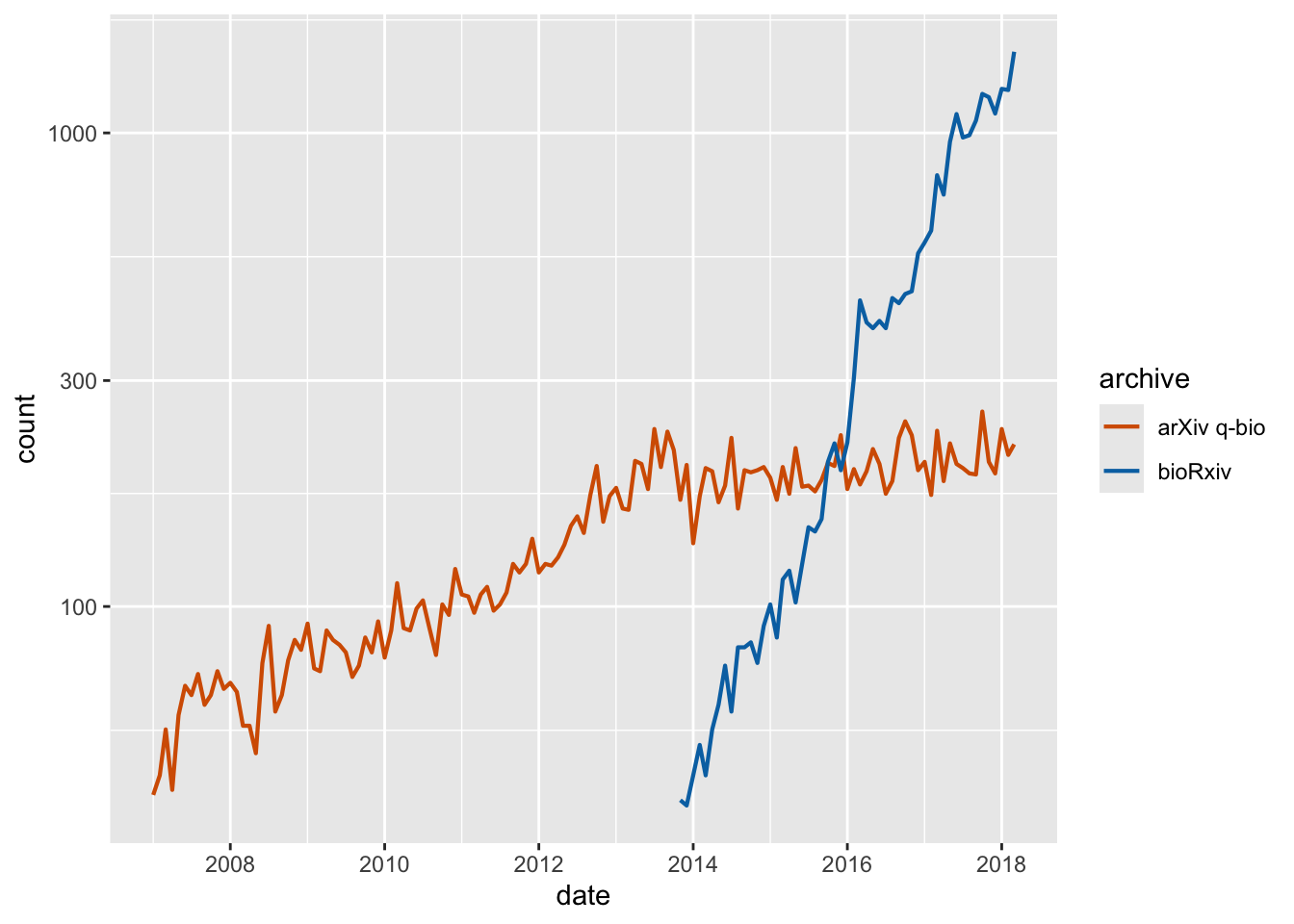
preprints_final <- filter(preprints, date == max(date))
ggplot(preprints) +
aes(date, count, color = archive) +
geom_line(linewidth = 0.75) +
scale_y_log10(
limits = c(29, 1600),
breaks = c(30, 100, 300, 1000),
expand = c(0, 0),
name = "preprints / month",
sec.axis = dup_axis(
breaks = preprints_final$count,
labels = preprints_final$archive,
name = NULL
)
) +
scale_x_date(name = "year", expand = c(0, 0)) +
scale_color_manual(values = c("#D55E00", "#0072B2"), guide = "none") +
theme_minimal_grid() +
theme(
axis.ticks.y.right = element_blank(),
axis.text.y.right = element_text(
size = 14,
margin = margin(0, 0, 0, 0)
)
)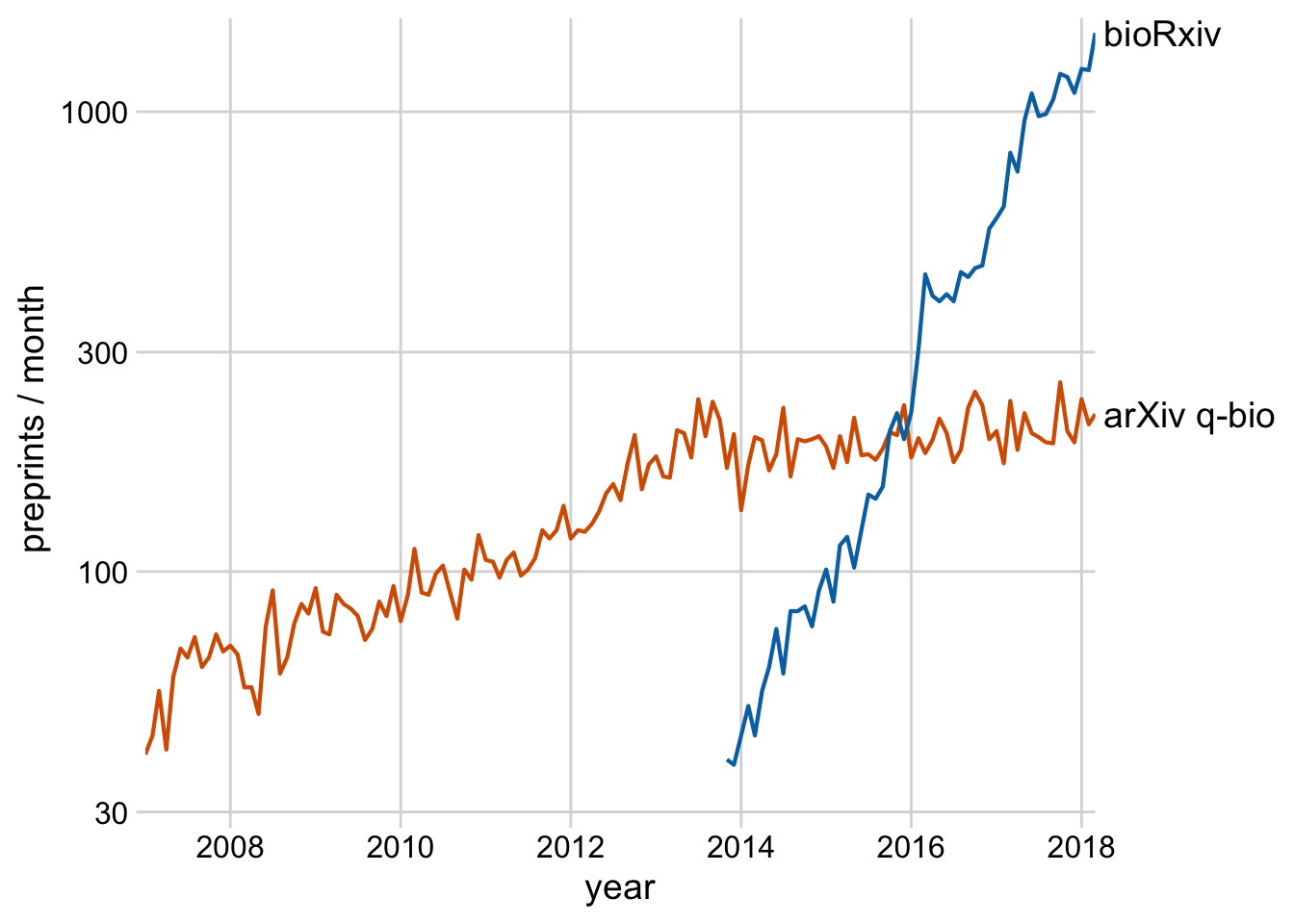
Exercise 1.2: Take the plot from Exercise 1.1 and label it with annotate() instead of using a secondary axis. Also color the text labels.
ggplot(preprints) +
aes(date, count, color = archive) +
geom_line(linewidth = 0.75) +
scale_y_log10(
limits = c(29, 1600),
breaks = c(30, 100, 300, 1000),
expand = c(0, 0),
name = "preprints / month"
) +
scale_x_date(name = "year", expand = c(0, 0)) +
scale_color_manual(values = c("#D55E00", "#0072B2"), guide = "none") +
annotate(
geom = "text",
label = c("arXiv q-bio", "bioRxiv"),
x = ymd(c("2012-05-01", "2016-12-01")),
y = c(180, 700),
hjust = c(1, 1),
color = colorspace::darken(c("#D55E00", "#0072B2"), .3),
size = 14,
size.unit = "pt"
) +
theme_minimal_grid()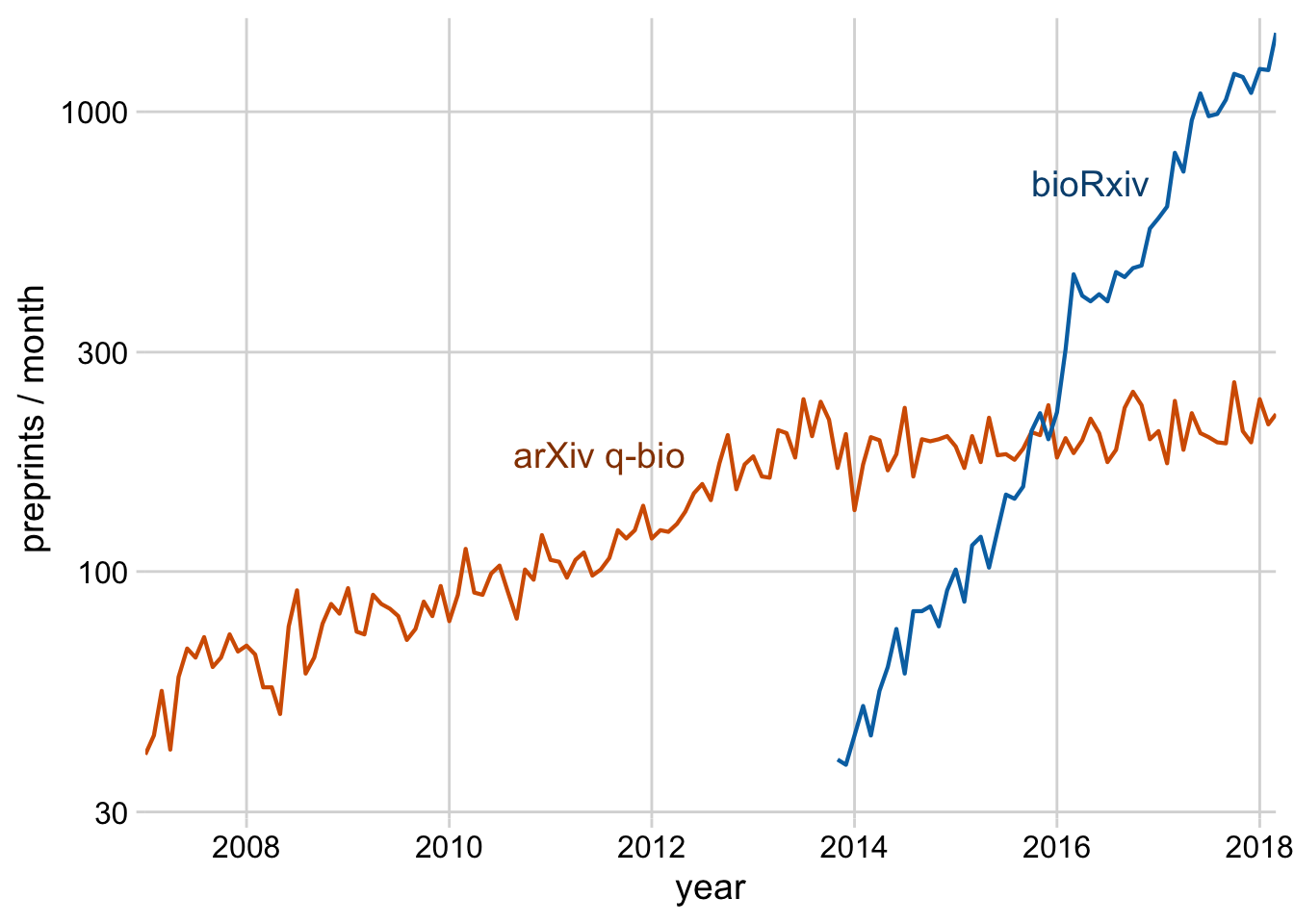
Exercise 1.3: Style this plot and place the legend in the correct order.
library(palmerpenguins)
penguins |>
na.omit() |>
ggplot() +
aes(body_mass_g, sex, fill = species) +
geom_boxplot(position = "dodge")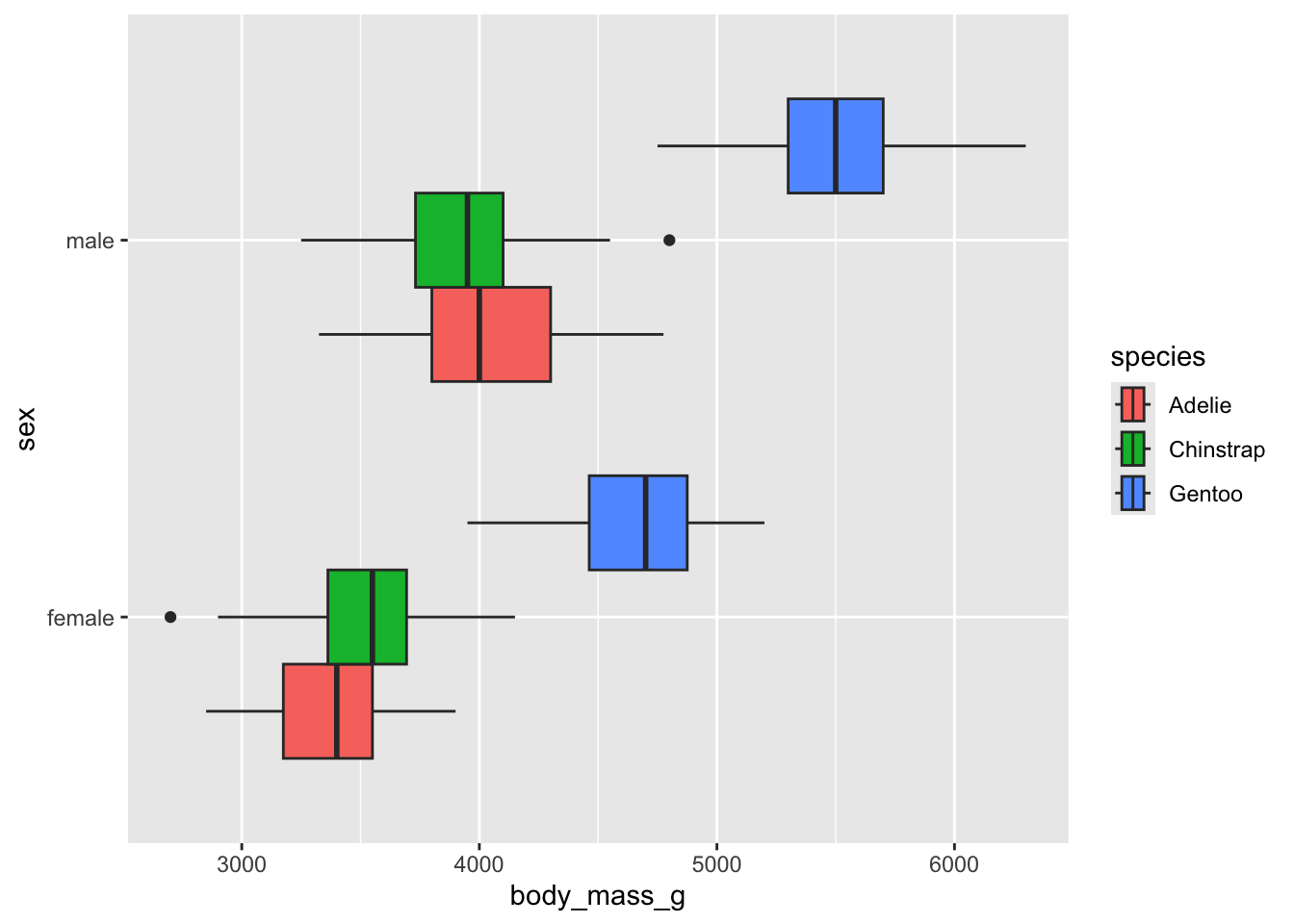
p <- penguins |>
na.omit() |>
ggplot() +
aes(body_mass_g, sex, fill = species) +
geom_boxplot(position = "dodge") +
scale_fill_manual(
values = c("#E69F00", "#56B4E9", "#009E73"),
name = NULL,
# when categorical variables are placed along the y axis, the legend
# generally has to be reversed
guide = guide_legend(reverse = TRUE)
) +
theme_minimal_vgrid() +
panel_border() +
theme(
legend.key.width = grid::unit(32, "pt")
)
p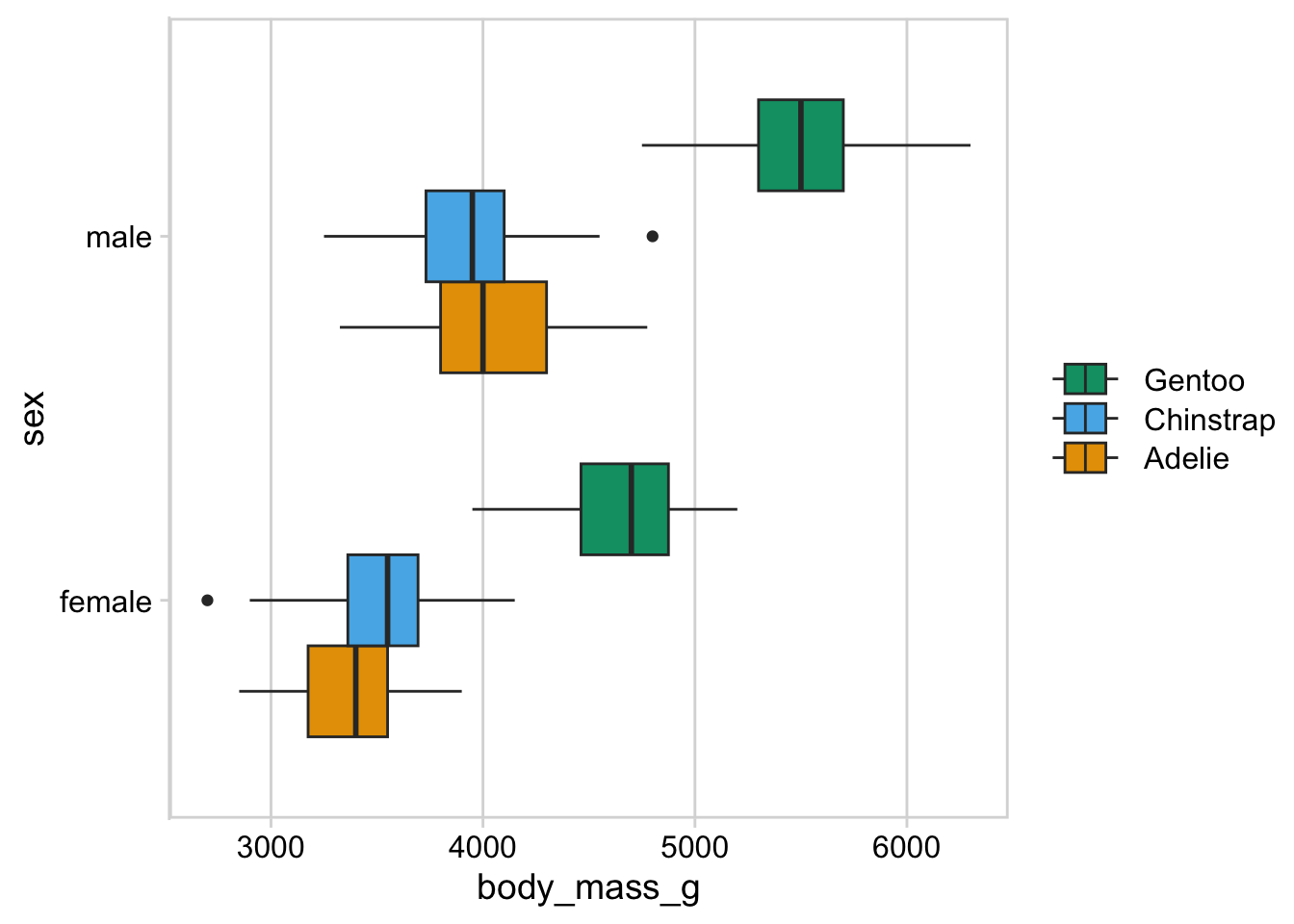
For this plot, a legend along the top of the plot looks better to me. You still want the ordering to match though (left-to-right should match top-to-bottom).
p + theme(legend.position = "top")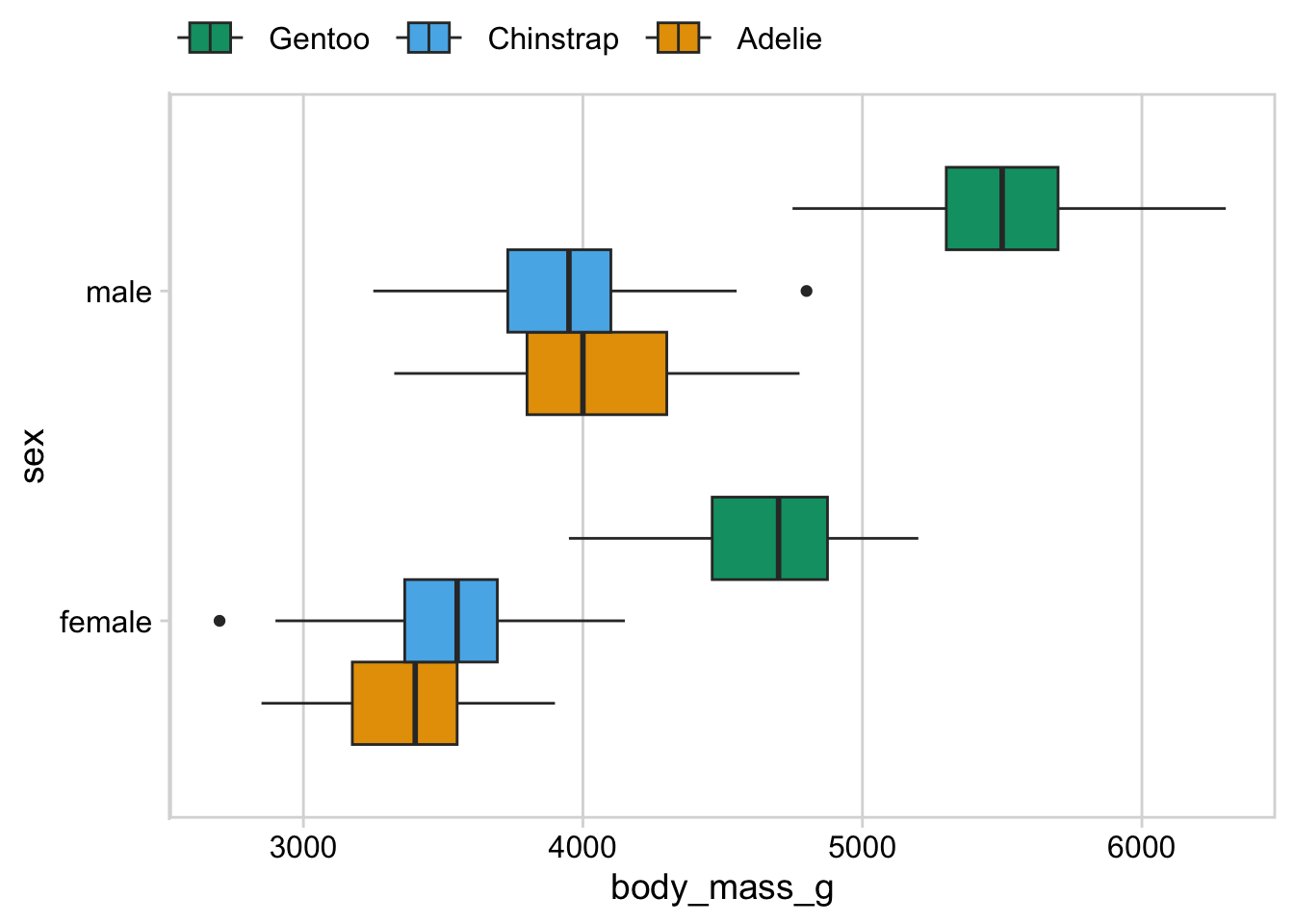
Solutions, Section 2
Exercise 2.1: Take the preprint plot from a prior exercise and move the legend inside the plot.
preprints <- read_csv("https://wilkelab.org/dataviz_shortcourse/datasets/preprints.csv") |>
filter(archive %in% c("bioRxiv", "arXiv q-bio")) |>
filter(count > 0)
ggplot(preprints) +
aes(date, count, color = archive) +
geom_line(linewidth = 0.75) +
scale_y_log10() +
scale_x_date() +
scale_color_manual(values = c("#D55E00", "#0072B2"))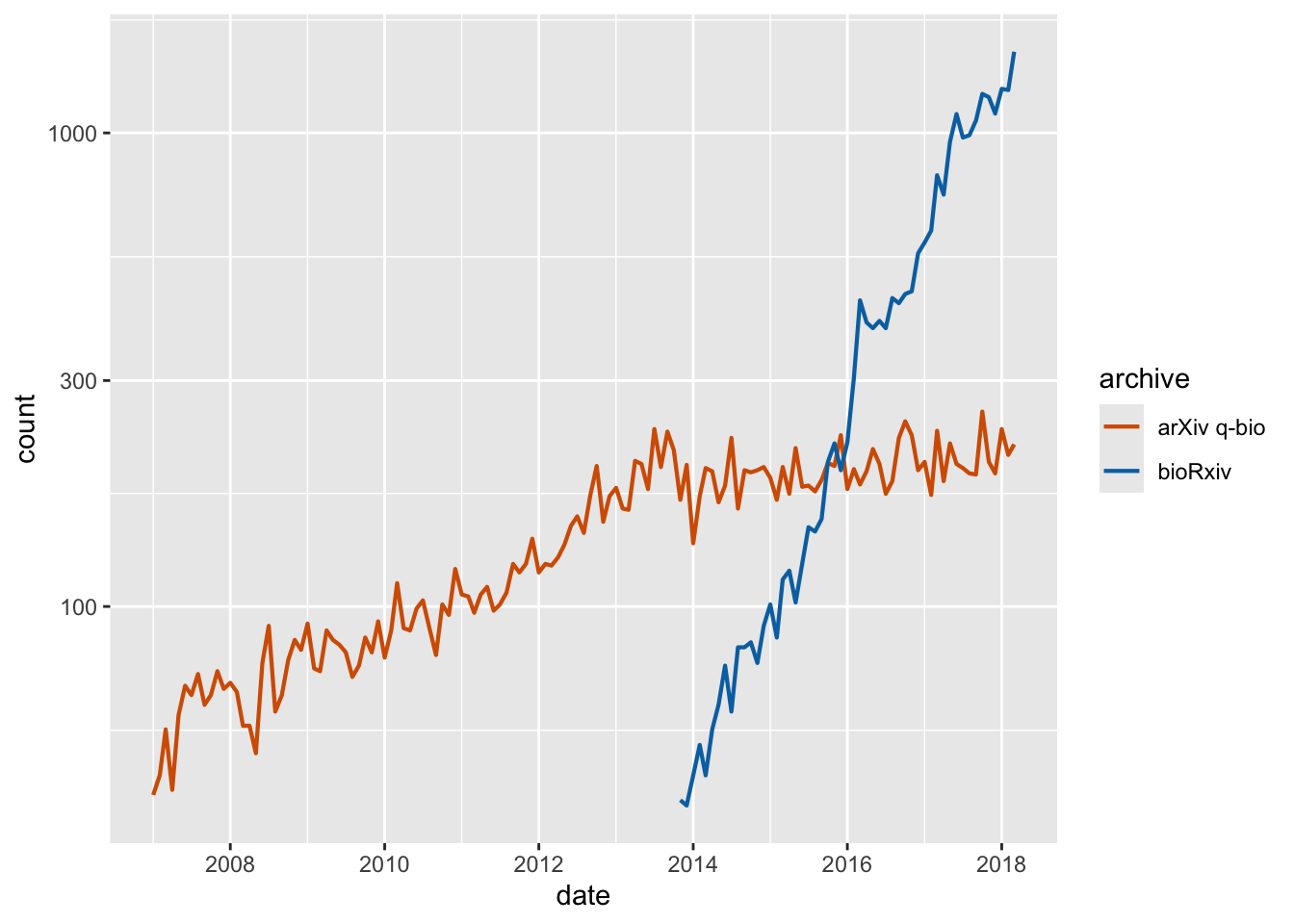
ggplot(preprints) +
aes(date, count, color = archive) +
geom_line(linewidth = 0.75) +
scale_y_log10() +
scale_x_date() +
scale_color_manual(
name = NULL,
values = c("#D55E00", "#0072B2"),
guide = guide_legend(reverse = TRUE)
) +
theme_minimal_grid() +
theme(
legend.position = "inside",
legend.position.inside = c(0.96, 0.1),
legend.justification = c(1, 0),
legend.box.background = element_rect(fill = "white", color = "white"),
legend.box.margin = margin(7, 7, 7, 7)
)